From Wikipedia, the free encyclopedia.
"Independent component analysis (ICA) is a computational method for separating a multivariate signal into additive subcomponents supposing the mutual statistical independence of the non-Gaussian source signals. It is a special case of blind source separation. When the independence assumption is correct, blind ICA separation of a mixed signal gives very good results. It is also used for signals that are not supposed to be generated by a mixing for analysis purposes. A simple application of ICA is the “cocktail party problem”, where the underlying speech signals are separated from a sample data consisting of people talking simultaneously in a room. Usually the problem is simplified by assuming no time delays or echoes. An important note to consider is that if N sources are present, at least N observations (e.g. microphones) are needed to get the original signals. This constitutes the square (J = D, where D is the input dimension of the data and J is the dimension of the model)."
The ICA Front End utility allows to perform the Independent Component Analysis using a publicly available software (by Sigurd Enghoff, Computational Neurobiology Lab) and storing the results of the analysis (a matrix) into a NPX file.
Then, you can load the computed weights by the NPX Lab software tools and ICA components can be easily visualized and all the NPX Lab Suite processing can be performed on these signals. In fact, it is possible to perform spectral and statistical analyses on sources, compute averages with the ERP viewer, etc… After the ICA Front End utility has stored ICA weights into a NPX File, the ICA processors is available on the Traces/Averages Processors Sidebar:
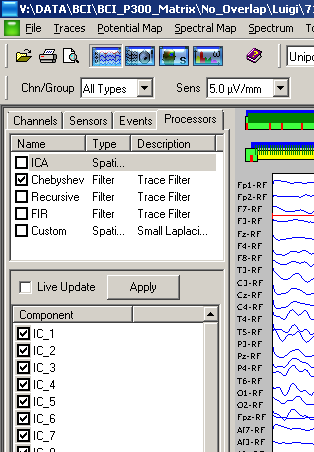
Note that initially its relative checkbox is deactivated, meaning that ICA components are not computed (they can be displayed, but they will appear as a null signal). ICA components are internally treated as if they were new acquired sensors (virtual sensors, meaning that they are not real sensors such as the EEG ones). The reason why into a NPX file are not stored the ICA components but just its weights is that the NPXLab tool allows you to selectively decide which component can be removed from your signals and instantaneously see how they change!
By double clicking on the ICA item in the same processors list you activate the ICA editor which allows you to see, component by component, how they are weighted over the scalp:
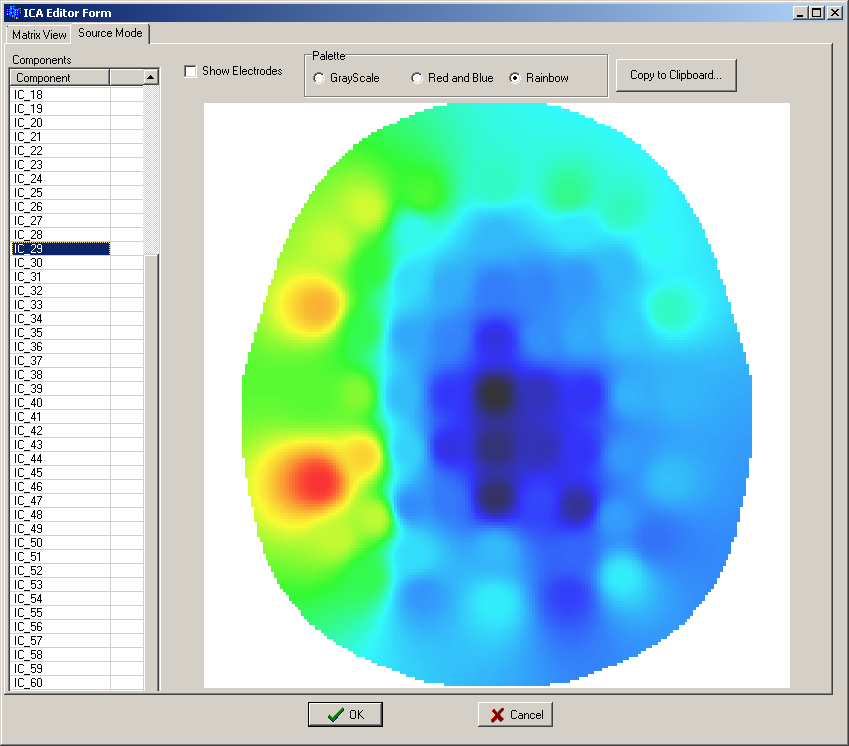
This is very useful to deduce the physiological or noise component they represent.
Usually one wants to remove some noise sources from the signals by means of the visual inspection of the components.
To see the ICA components, you have to:
1) check the ICA checkbox among the processors list. This will enable the computation of the ICA components from the original acquired signals.
2) select the components (again, check the checkbox) that you want to visualize from the channel sensors sidebar
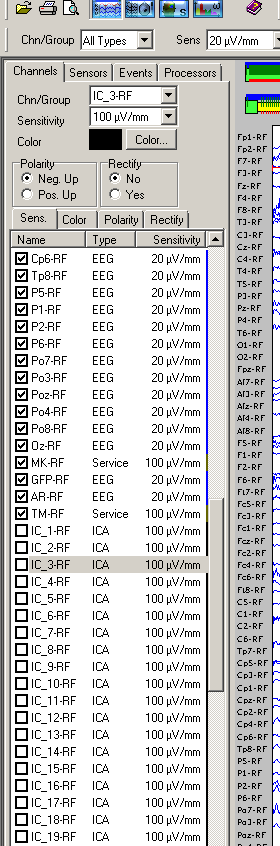
Now you can visualize the ICA components and decide which one to include/exclude from the acquired signals.
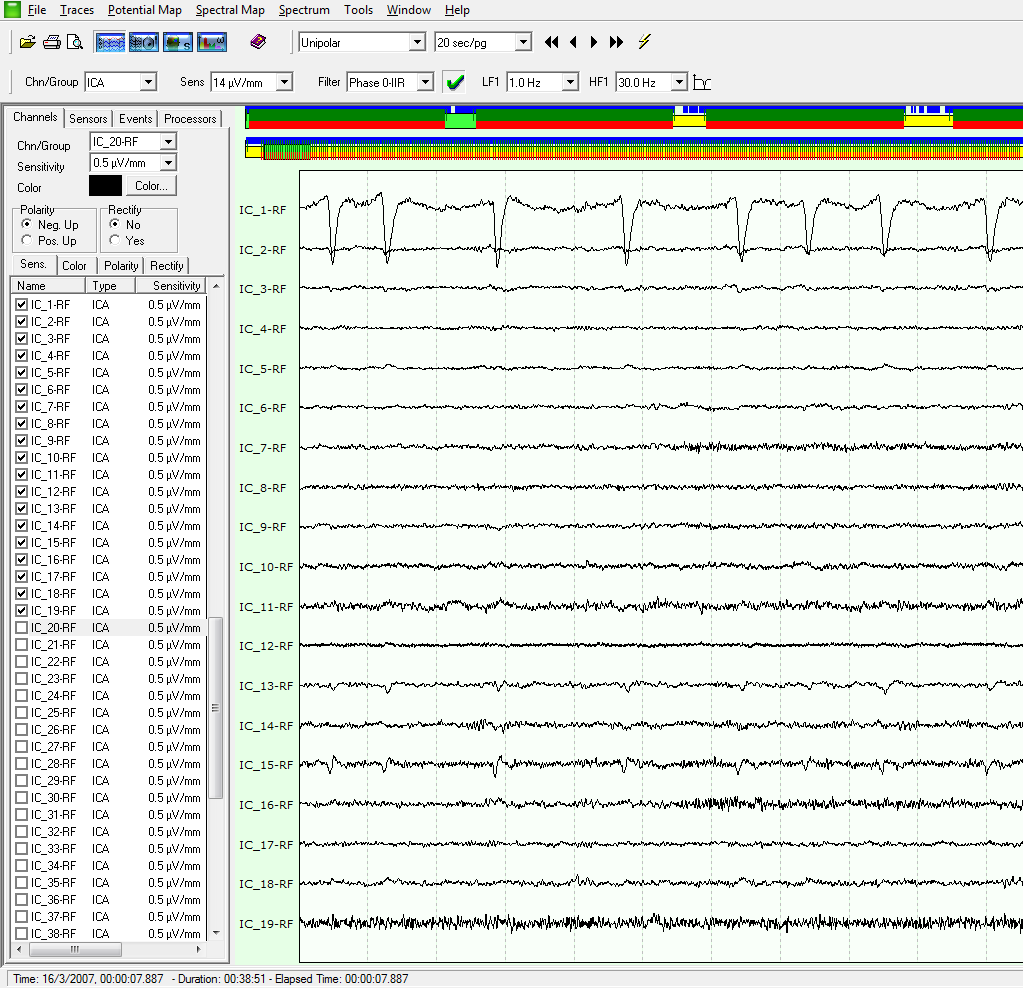
ICA sources can be “removed” from EEG data: once one has identified a noise source (e.g. eye movements) it can be subtracted from the EEG data, thus cleaning them. In the following figures it is shown the effect on the EEG (blue) traces when the IC_1 component, which appears to be related to EOG activity, is switched off (checkbox was unchecked).
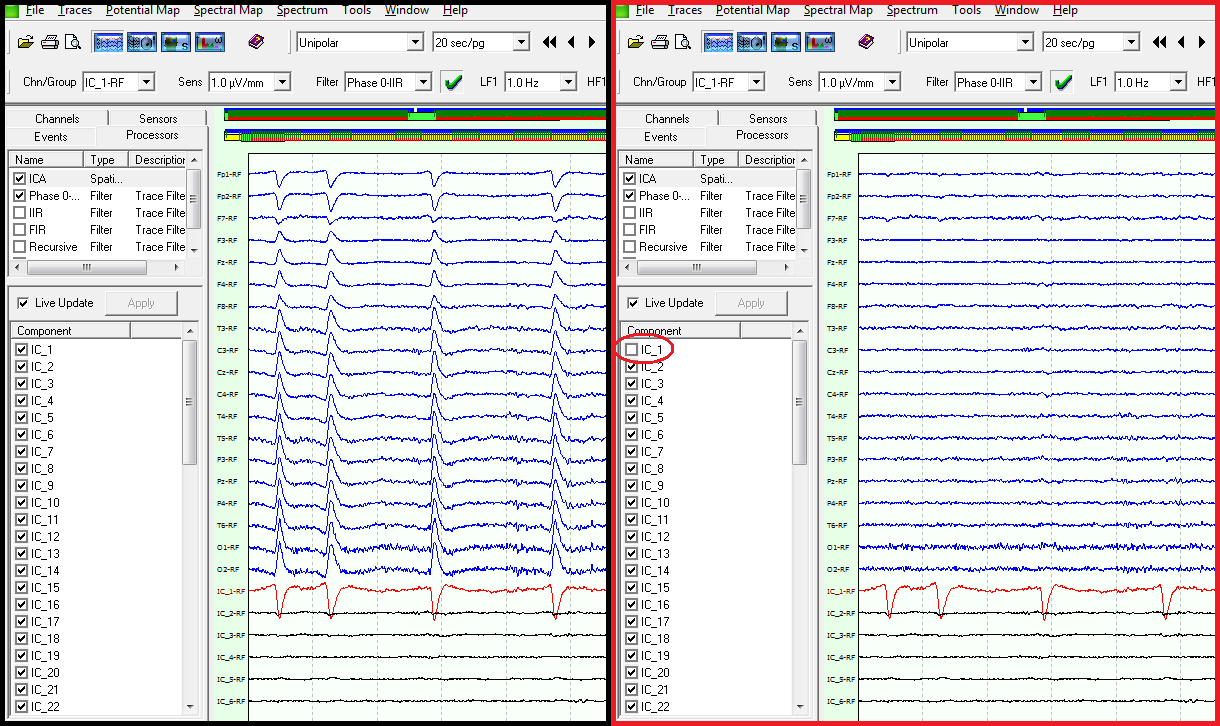
This powerful operating modality, which is unavailable even in most commercial tools, allows to quickly increase the signal to noise ratio of you data, or to analyze just an ICA component that is related to some specific brain activity (e.g. you may detect a P300 component and compute the spectral and statistical maps of it!).
Created with the Personal Edition of HelpNDoc: Full featured Documentation generator